Naveen Duhan
Building AI-driven bioinformatics systems that translate complex biological data into clear, actionable insight.
I collaborate with experimental and computational teams to turn noisy sequencing data into clear, decision-ready biological insight.
Open to collaborations in multi-omics, host–pathogen systems, and AI for genomics.

Across academia and applied research, I have led projects that span NGS analysis, multi-omics integration, and the development of web resources that make complex data accessible. I enjoy collaborating with interdisciplinary teams to move from data to decisions.
Where computation meets biology
A snapshot of the research themes and technologies guiding my work in genomics, systems biology, and AI.
Multi-omics Data Integration
Developing computational methods to merge genomics, transcriptomics, proteomics, and metabolomics for a systems-level view of biology.
AI in Genomics
Applying deep learning to predict gene function, regulatory elements, and variant effects with interpretable, robust models.
Systems Biology
Modeling biological networks and pathways to uncover mechanism-level insights and identify intervention points.
Bioinformatics Tooling
Designing databases, web servers, and analysis pipelines that make complex data usable for researchers.
Tools and platforms built for researchers
A selection of computational tools, databases, and web servers created to make complex biological data accessible.
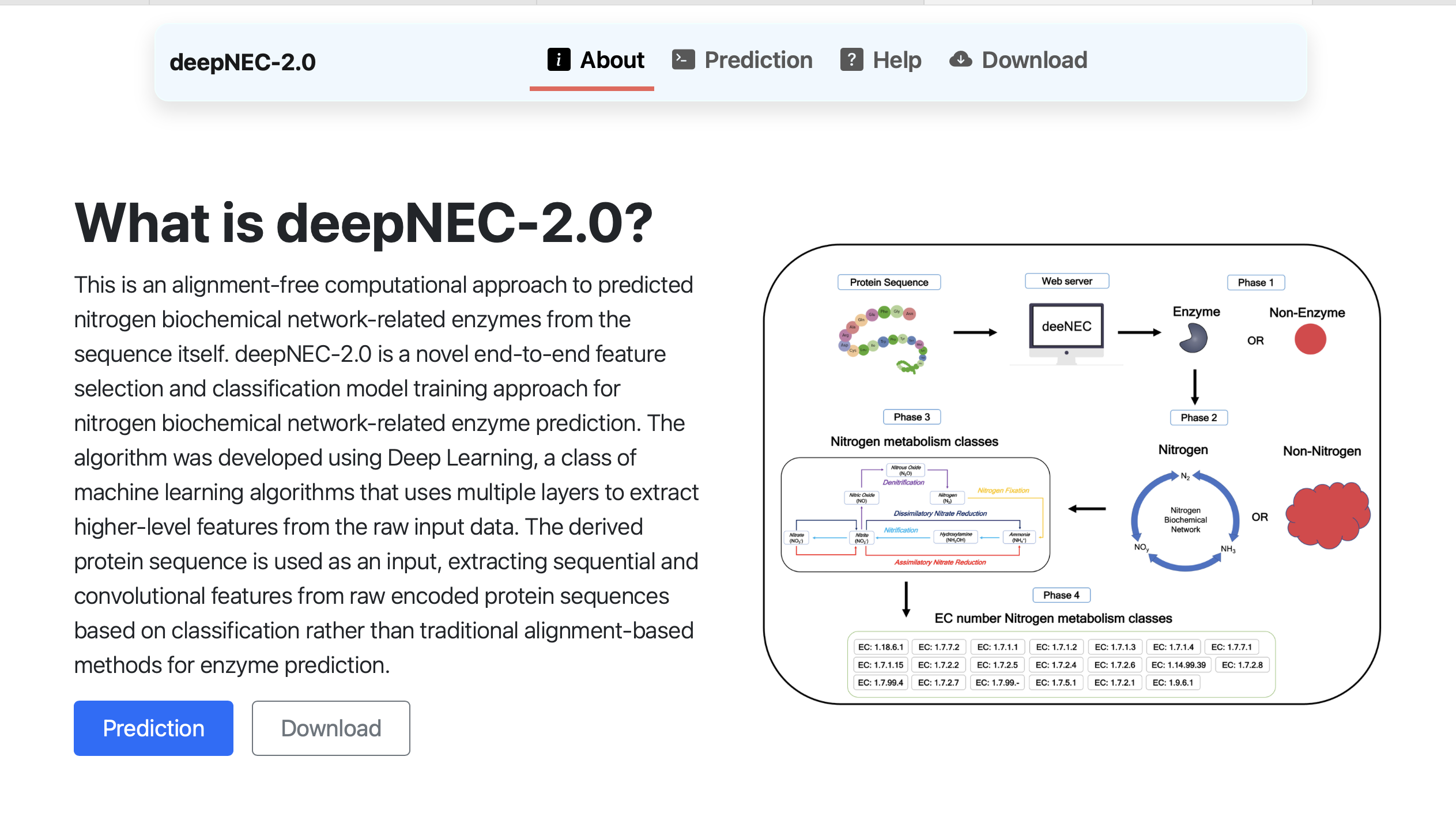
deepNEC
A deep learning framework for predicting nitrogen metabolism enzymes from protein sequences, providing interpretable insights into sequence-function relationships.
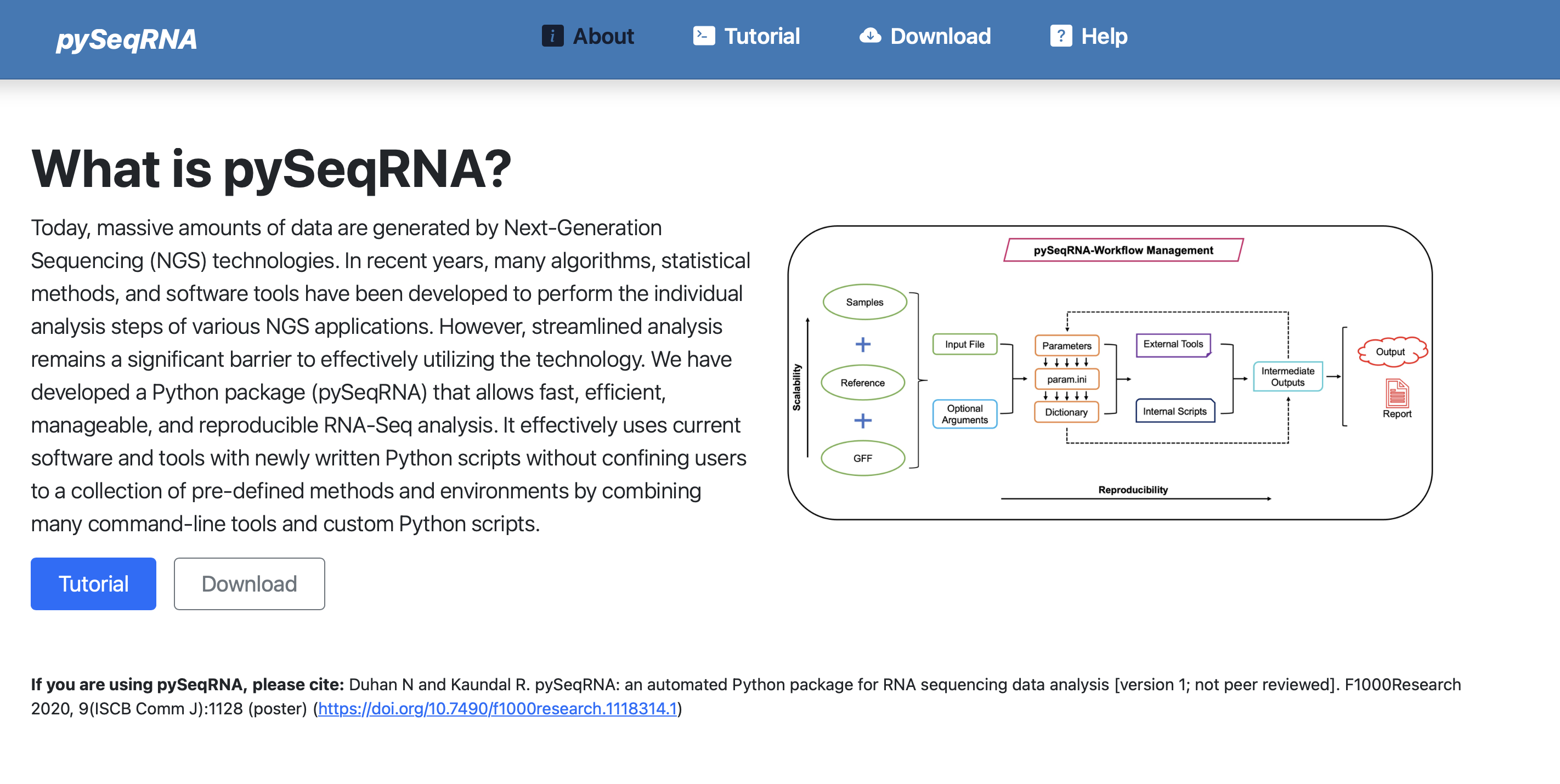
pySeqRNA
A comprehensive RNA-Seq analysis package with automated QC, alignment, quantification, and differential expression workflows.
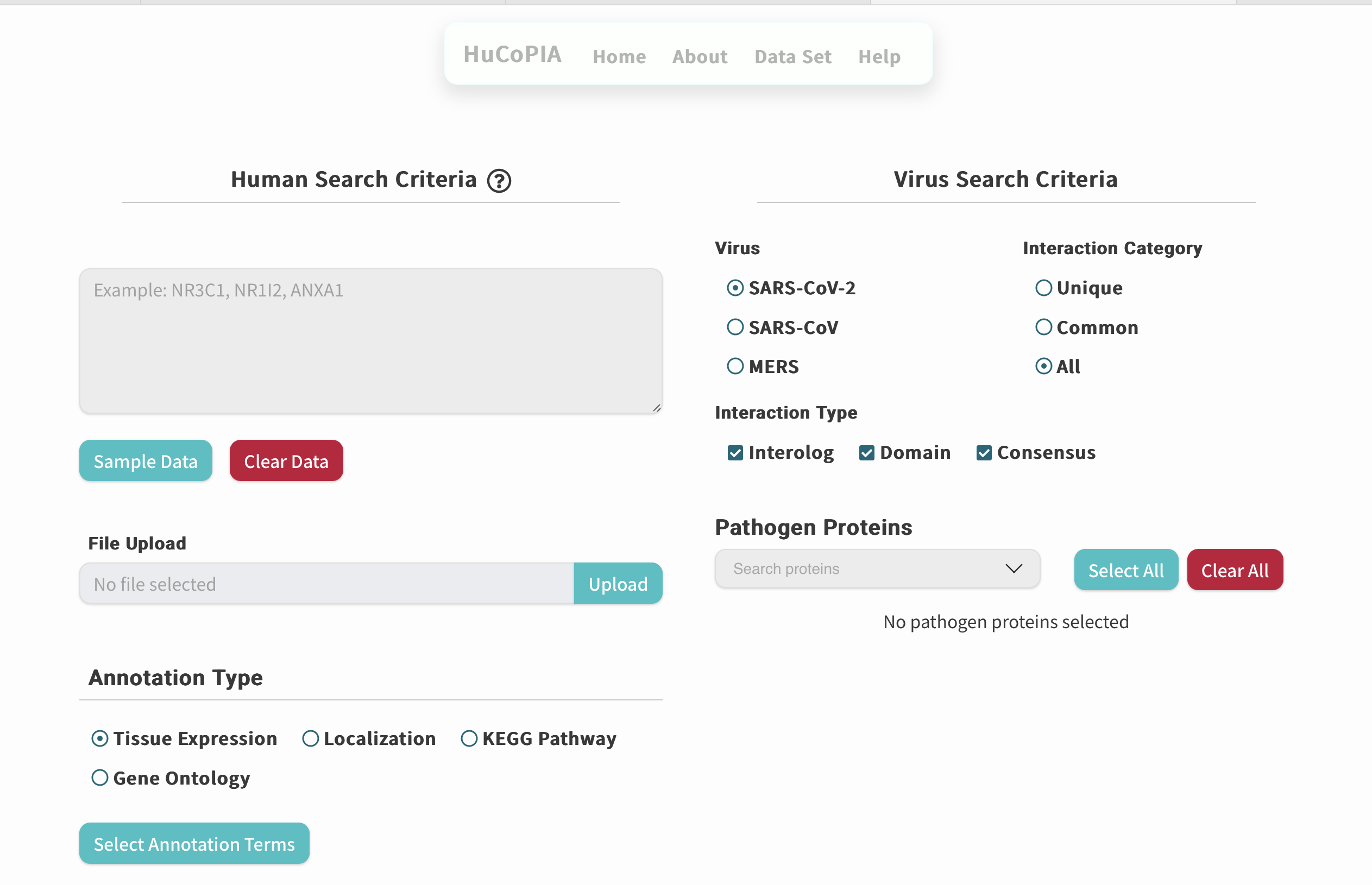
HuCoPIA
An atlas of human-coronavirus protein interactions with network visualization and comparative analyses across Coronaviridae.
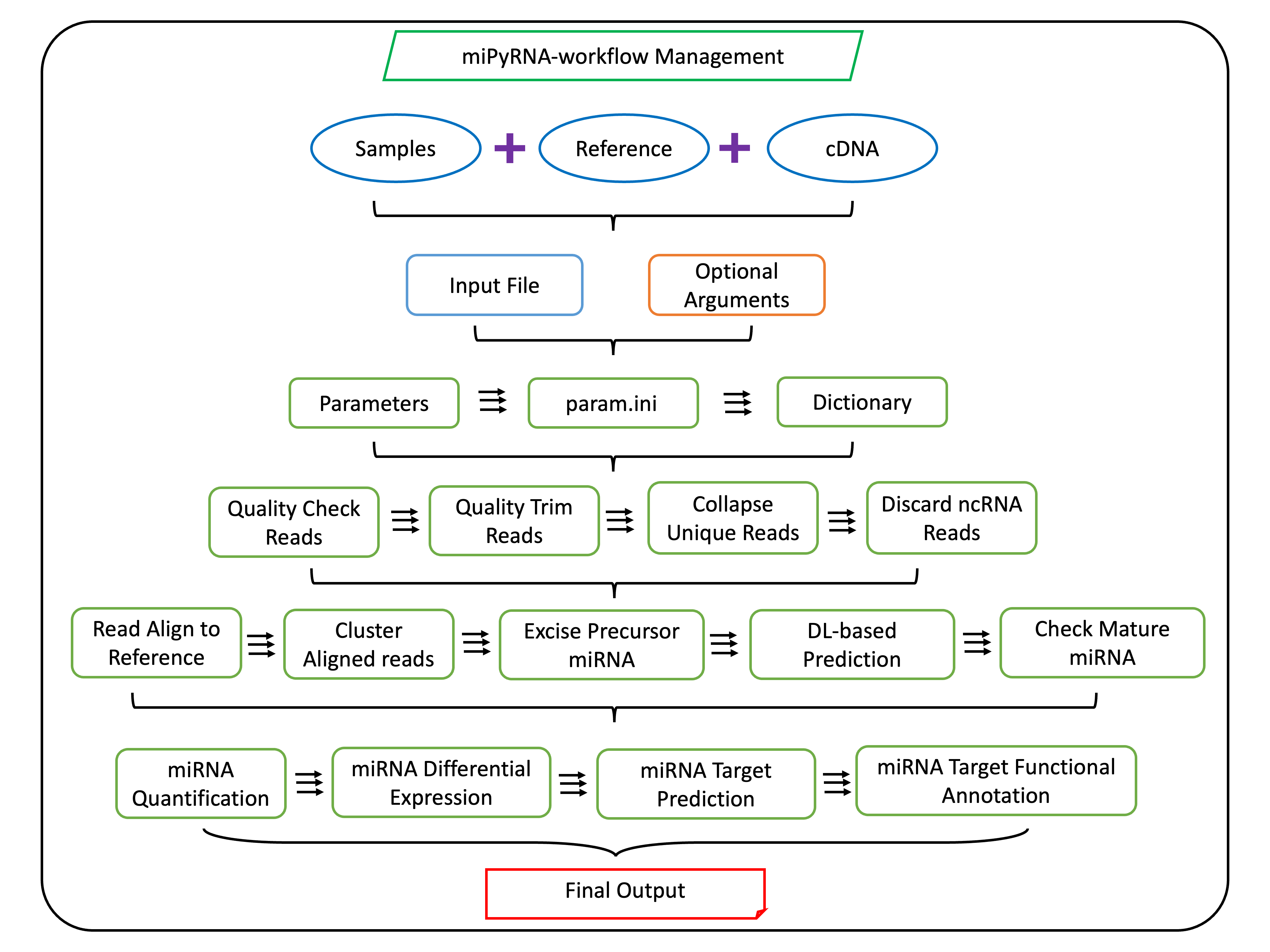
miPyRNA
A Python package for small RNA-seq analysis focused on microRNA identification, quantification, and target prediction.
Recent peer-reviewed work
Selected publications from computational genomics, host-pathogen interactions, and AI-enabled biology.
Discovery of potential haplotypes associated with varying levels of vicine content due to the InDel1.4 and a coding-SNP in the VC1 gene in faba bean (Vicia faba L.)
Sadhan Debnath and Wricha Tyagi and Mayank Rai and Kuldeep Singh and Sujan Majumder and Duhan, N. and Ng Tombisana Meetei
Plant Gene • 41 • 100481 (2025)
Comparative miRNome and transcriptome analyses reveal the expression of novel miRNAs in the panicle of rice implicated in sustained agronomic performance under terminal drought stress
Kaur, S. and Seem, K and Duhan, N. and Kumar, S and Kaundal, R and Mohapatra, T
Planta • 259 • 128 (2024)
RSLpred2: An integrated web server for the annotation of Rice proteome subcellular localization using deep learning
Duhan, N. and Kaundal, R.
Rice • Accepted (2024)
Geographical Expansion of Avian Metapneumovirus Subtype B: First Detection and Molecular Characterization of Avian Metapneumovirus Subtype B in US Poultry
Luqman, M. and Duhan, N. and Temeeyasen, G. and Selim, M. and Jangra, S. and Mor, S.K.
Viruses • 16 • 508 (2024)
Training the next generation of bioinformaticians
Coursework, workshops, and mentorship across US and international programs.
Software, databases, and web servers
A complete catalog of software tools and research web resources developed or maintained.
Packages supporting RNA analysis, variant detection, and deep learning.
Databases and web servers covering host-pathogen interactions and genomic markers.
Notes from the lab
Insights on reproducible bioinformatics, AI in genomics, and multi-omics strategy.
Designing Reproducible Bioinformatics Pipelines
A practical framework for building pipelines that are transparent, testable, and resilient to data drift.
Deep Learning for Genomics: What Works in Practice
Lessons learned from deploying deep learning models for sequence analysis and functional prediction.
From Sequences to Systems: A Multi-omics Perspective
Why integrated omics is crucial for understanding complex traits and host-pathogen interactions.